Overview
Revisitting the sparse matrix technology.
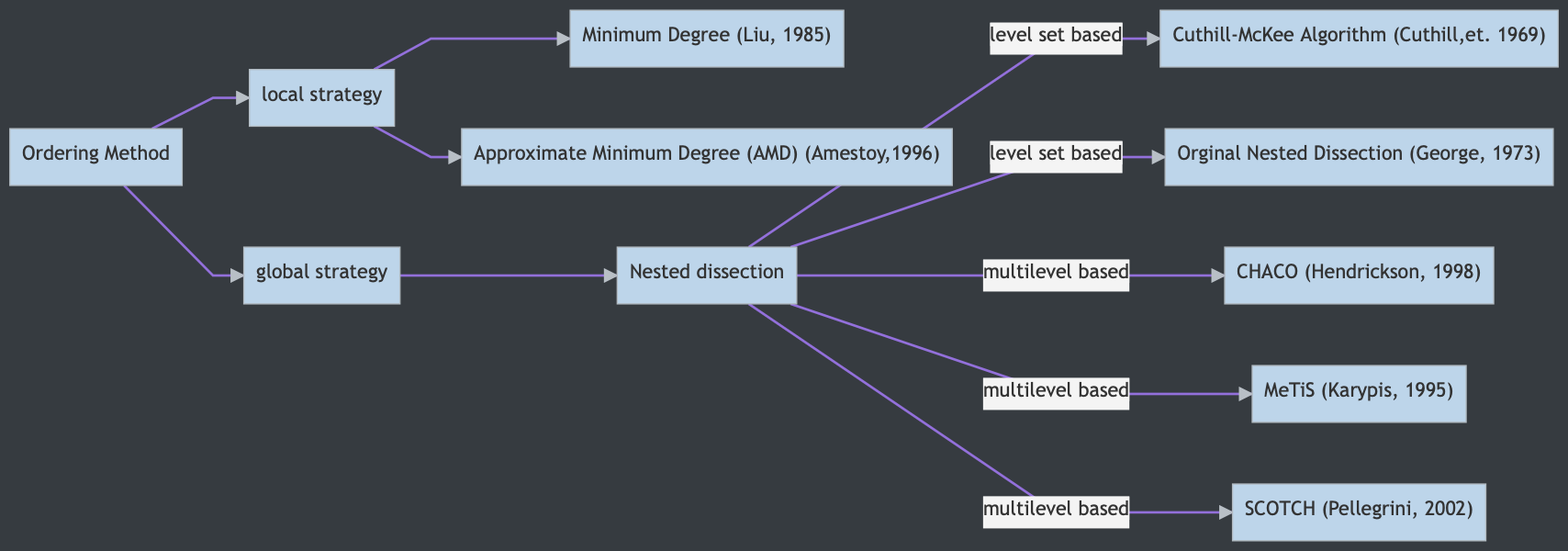
Level set based Nested Dissection (George, 1973)
1. Essential terminology
Given undirected graph $G(V,E)$:
- The Path is an ordered set of distinct vertices
- Eg: A path of length K = ${v_1,v_2,\cdots,v_k,v_{k+1}}$
- The Distance between two vertices u and v: $dist(u,v)$ is the length of the shorest path
- $dist(u,v)$ = shorest path
- The Diameter of graph $G$ is the maximum distance between ANY two vertices in the graph:
- $diam(G)=max_{u,v\in G}\ dist(u,v)$
- A vertex $v$ is Extremal with respect to a vertex $u$ if v is as far away from u as possible:
- $dist(u,v) = max_{w}\ dist(u,w)$
- When two vertices u and v are mutually extremal <=> They form a pseudo-diameter pair <=> u and v are both pseudo-peripheral vertices:
- $dist(u,v)=max_w\ dist(u,w) = max_v\ dist(v,w)$
- Level sets: GIVEN ANY VERTEX $s$ FROM GRAPH:
- One level set is the collection of vertices with the same distance from $s$:
- $v\in L_i(s)$ if and only if $dist(v,s)=i$
- The Level sets of a root vertex $s$:
- $L(s)={L_0(s), L_1(s),\cdots,L_k(s)}$
- One level set is the collection of vertices with the same distance from $s$:
2. Vanilla Nested Dissection
2.1 Some useful conclusions
FACT 1 Each level set defines a separator
- An edge occurs only when two vertices belong to the same level set or to adjacent level sets
FACT 2 A good seperator should be as small as possible
- Small separators cause the shaded areas associated with the S-sets to be small
FACT 3 Using a pseudo-peripheral vertex as the root is more likely to find a good seperator.
- A level structure rooted at a pseudoperopheral vertex is likely to have many levels
2.2 Algorithm
Phase 1: Finding the pseudoperopheral vertices (Mutual extremal)
Code
def diameter_pair_algo(G) -> set:
"""
find one pseudo-diameter pair
: param G: Graph
:return: a pair of mutual-extremal vertexes (s,t)
"""
s = np.random.choice(G.nodes)
while True:
level_set = level_set_algo(G,s)
# find th extremal w.r.t root
t = np.random.choice(level_set[-1])
level_set_n = level_set_algo(G,t)
if len(level_set) == len(level_set_n):
return (s,t)
s = t
Phase 2: Constructing the level sets
Code:
def level_set_algo(G, root) -> list:
"""
find the level set with respect to a root vertex
:param root: root vertex, the starting point
:return: level set, ordered by distance
"""
pathes = nx.single_source_shortest_path(G,root)
pathes_length = [len(route) - 1 for route in pathes.values()]
pathes_nodes = list(pathes.keys())
level_set = []
for i, vertex in enumerate(pathes_nodes):
if pathes_length[i] == pathes_length[i-1]:
level_set[-1].append(vertex)
else:
level_set.append([vertex])
if sum([len(i) for i in level_set]) != len(G.nodes):
nx.draw(G, with_labels=True)
plt.savefig('test.png')
print("bla")
return level_set
**Phase 3: Find the inital seperator **
Code
def single_partition(G, ALPHA=4.0) -> dict:
"""
single layer level set based partition algorithm
The emhanced method of GeorgeLiu Algo with cost function (Ashcraft, 2016)
:return: the partition result, grouped by Black, White and Seperator
"""
s,_ = diameter_pair_algo(G)
level_set = level_set_algo(G,s)
cost = np.Inf
partition = {}
for i in range(1,len(level_set)-1):
# construct partition set
B = level_set[:i]
W = level_set[i+1:]
S = level_set[i]
cur_cost = cost_1(B,W,S, ALPHA)
if cur_cost < cost:
partition["B"] = B
partition["W"] = W
partition["S"] = S
cost = cur_cost
# when the len(level_set) < 3:
if len(partition) < 3:
assert len(partition) < 2, "check"
partition["B"] = [level_set[0]]
partition["S"] = level_set[1]
partition["W"] = [[]]
return partition
return partition
**Phase 4: Recursive excecution **
Code
def ND_solve(G,p) -> list:
"""
main function to excecute the partition recursively
:param p: the list of reordered vertex index
:return: the reordered index of vertexes
"""
# filtering the isolated vertex
iso_nodes = singleton(G)
p += iso_nodes
for node in iso_nodes:
G.remove_node(node)
if len(list(G.nodes)) <= 2:
p += list(G.nodes)
return p
partition = single_partition(G)
B = [item for sublist in partition["B"] for item in sublist]
W = [item for sublist in partition["W"] for item in sublist]
seperator = partition["S"]
sub_graphs = [G.subgraph(c).copy() for c in [W,B]]
ND_solve(sub_graphs[0],p)
ND_solve(sub_graphs[1],p)
p += seperator
return p